Epigenetic Analysis
Overview
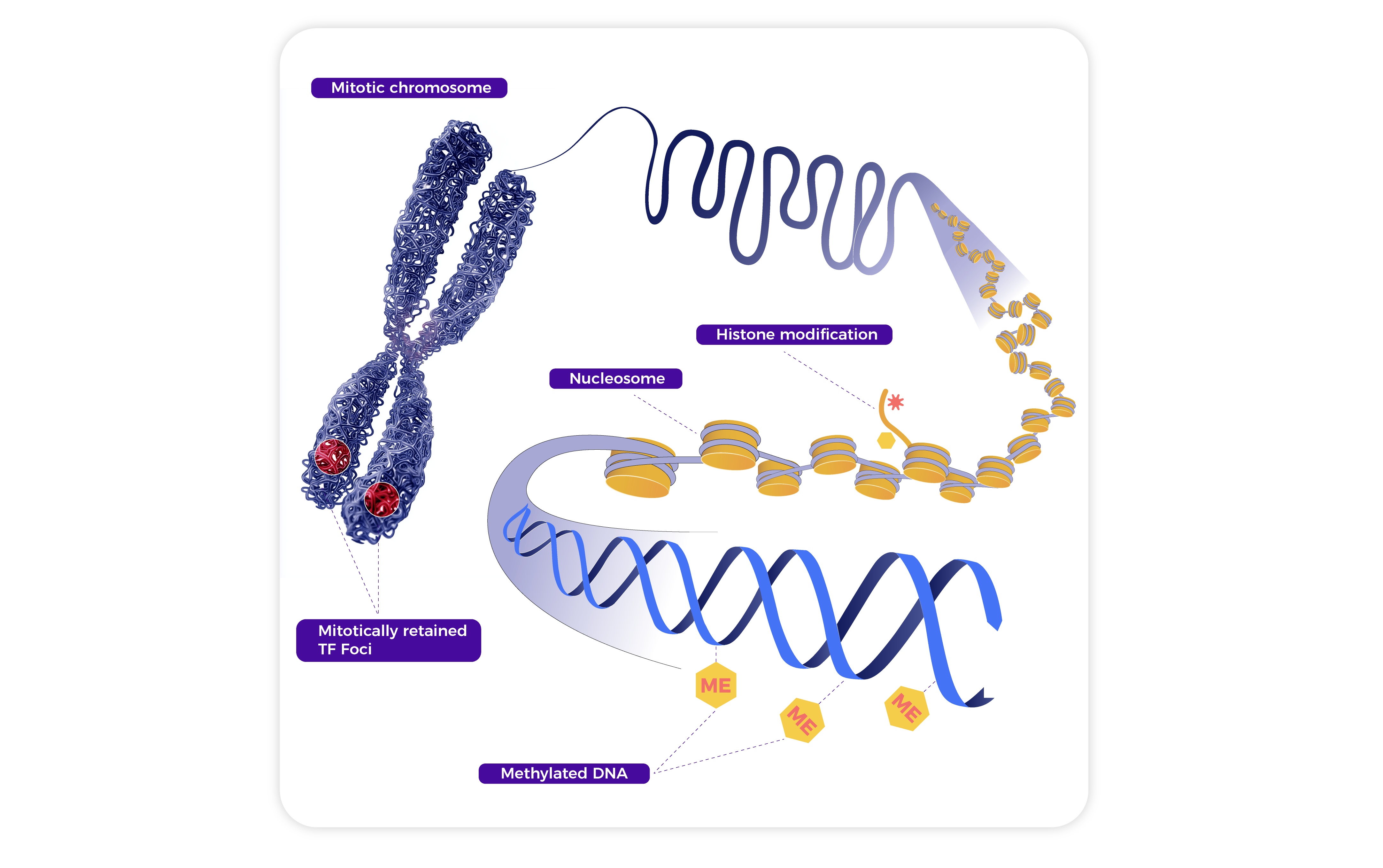
Epigenetics investigates heritable changes in gene expression that occur without alterations to the underlying DNA sequence. It plays a pivotal role in development, cellular differentiation, and responses to environmental signals.
Two primary mechanisms underlie epigenetic regulation:
1. DNA Methylation: Involves the addition of methyl groups to cytosines, typically at CpG sites, leading to gene silencing. It regulates genomic imprinting, X-inactivation, and transposon suppression, and is often dysregulated in diseases such as cancer.
2. Histone Modifications: Chemical changes to histone proteins (e.g., acetylation, methylation) alter chromatin structure and influence transcription. These modifications serve as signals for activating or repressing gene expression.
To investigate these mechanisms, several advanced techniques are employed:
1. Bisulfite Sequencing: Enables single-base resolution mapping of DNA methylation by converting unmethylated cytosines to uracil, preserving methylated ones. Used in both genome-wide and targeted studies.
2. ChIP (Chromatin Immunoprecipitation): Identifies DNA regions bound by specific proteins or carrying histone modifications. When combined with sequencing (ChIP-seq), it maps regulatory elements and chromatin states.
3. ATAC-seq: A fast and sensitive method to profile open chromatin regions, revealing active regulatory elements such as promoters and enhancers,even with minimal input material.
Together, these tools provide critical insights into how epigenetic regulation shapes gene expression and contributes to health and disease.





